-
Publish Your Research/Review Articles in our High Quality Journal for just USD $99*+Taxes( *T&C Apply)
Offer Ends On
Jola Rahmahani*, Arshita Buwana Dwiandari, Endang Suprihati, Suwarno, Hani Plumeriastuti, Aswin Rafif Khairullah and Fedik Abdul Rantam
Corresponding Author: Jola Rahmahani, Department of Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Jl. Dr. Ir. H. Soekarno, Kampus C Mulyorejo, Surabaya 60115, East Java, Indonesia.
Received: September 09, 2022 ; Revised: October 15, 2022 ; Accepted: October 18, 2022 ; Available Online: November 08, 2022
Citation: Rahmahani J, Dwiandari AB, Suprihati E, Suwarno, Plumeriastuti H, et al. (2022) Nucleotide, Homology and Phylogenetic Analyses of Gene Encodes Hexon Protein of Fowl Adenovirus from Broiler Chicken with Inclusion Bodies Hepatitis. J Vet Marine Res, 2(1): 1-8.
Copyrights: ©2022 Rahmahani J, Dwiandari AB, Suprihati E, Suwarno, Plumeriastuti H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Views & Citations
Likes & Shares
Inclusion Bodies Hepatitis is caused by infection with FAdVs (Fowl adenovirus) of the Aviadenovirus genus. The distribution of IBH (Inclusion Bodies Hepatitis) in the world is increasing very high in poultry production. The IBH outbreak in Indonesia is included in the re-emerging disease, which means that although it has begun to fade, it does not rule out the possibility that it will reappear. This study was conducted to detect and analyze the characterization of FAdVs that attack broiler chickens in Salatiga based on homology and phylogenetic results. The sample consisted of three chickens suspected of being infected with FAdvs. They were collected from broiler farms that had no previous IBH vaccination record in Salatiga. Chicken livers were respectively used for molecular detection of the partial Hexon gene. Furthermore, molecular analyzes such as mutation, homology and phylogenetic analysis were carried out to study the molecular characteristics compared to the known sequences of FAdvs submitted in Genbank. Pathological features of broiler liver organs suspected of being infected with FAdVs showed that the liver looked slightly swollen, striped, there were areas of pale, fatty, and brittle necrosis. Among the three samples, one sample was positive for the Hexon gene FAdv genomic material (IBH/CHK/H3/SLTG/2020). Homology analysis showed that IBH/CHK/H3/SLTG/2020 had a 97% similarity with reference sequences and isolates from China. The results of nucleotide alignment sequencing showed a shift in the nucleotide sequence. In conclusion, the IBH/CHK/H3/SLTG/2020 samples have a fairly high similarity with various isolates from China. Phylogenetic analysis showed that the sample IBH/CHK/H3/SLTG/2020 was in the same clade with isolates from China and Japan but had a different clade with isolates from Germany.
Keywords: Inclusion Bodies Hepatitis, Fowl Adeno virus, PCR, Sequencing, Molecular analysis
INTRODUCTION
Inclusion Bodies Hepatitis is caused by infection with FAdVs (Fowl adenovirus) of the Aviadeno virus genus [1]. Aviadeno viruses are grouped into five species based on molecular structure and subdivided into 12 serotypes based on cross-neutralization tests, including fowl adenovirus A(FAdVs-1), fowl adenovirus B(FAdVs-5), fowl adenovirus C (FAdVs-4 and -10), fowl adenovirus D (FAdVs-2,-3,-9 and -11), and fowl adenovirus E (FAdVs-6,-7,-8a and -8b) [2].
The distribution of IBH (Inclusion Bodies Hepatitis) in the world is increasing very high in poultry production [3]. Every year, the number of cases of adenovirus infection in poultry increases, but the role of the virus in the pathogenesis is still unclear [4].The death rate from this disease reaches more than 7% and usually death occurs suddenly without showing clear clinical symptoms [5].
The characteristics of the main protein are widely used to determine the virulence of the gene in FAdVs isolates [6]. Molecular analysis of FAdVs showed that the adenovirus capsid is composed of three main protein structures, namely hexon, penton and fiber [7]. Molecular tests such as the Polymerase Chain Reaction followed by sequencing and phylogenetic studies have been carried out for the detection and differentiation of FAdVs [8].
In Indonesia, this disease first appeared in 1985 on the island of Java, namely Semarang Regency and DKI Jakarta [9]. This disease had faded, but at the end of 2017 to 2018, IBH infections were found in samples taken from cases in the field in the form of formalinated organs and fresh organs [10]. IBH outbreaks in Indonesia are included in the re-emerging disease, which means that although it is currently starting to fade, it does not rule out the possibility that it will reappear; causing an emergency situation in livestock and also the vaccine for IBH in Indonesia is still not available [11].
This study was conducted to detect and analyze the characterization of FAdVs that attack broiler chickens in Salatiga based on homology and phylogenetic results. Salatiga is one of the areas with a large number of livestock sectors but is still included in the fourth sector of broiler chicken farms, thus increasing the risk of an outbreak of IBH disease.
METHODS
Sample collection
This study used 3 broiler chickens that showed symptoms of IBH. After the chicken was necropsied, the liver was taken and put into an organ pot that had been given 10 ml of PZ physiological solution and added 0.1 ml of penicillin-streptomycin antibiotics each and then put into a coolbox to be brought to the laboratory. Samples were taken from broiler farms in Salatiga. This research was conducted at the Laboratory of Virology and Immunology, Department of Veterinary Microbiology, Faculty of Veterinary Medicine, Airlangga University, Surabaya. The time of the study was carried out in October - November 2020.
Organ suspension
Samples that were frozen were thawed first and then chopped using a blade and placed in a mortar and mortar while 10 ml of PZ physiological solution and penicillin-streptomycin antibiotics were added to be further ground until smooth with the maximum number of crushed organs being 25 mg. The results of the scouring of the organ samples were transferred to a centrifuge tube, and then centrifuged for 15 min at a speed of 6000 rpm. The centrifuged supernatant was taken using a syringe and transferred to a bar tube and added to the antibiotic Penicillin-Streptomycin.
Detection hexon gene
The DNA extraction process was carried out according to the QIAamp DNA Mini Kit Protocol [12]. The DNA amplification process was in accordance with the following program: the pre-denaturation process was set at 94⁰C for 10 min, the denaturation process at 94⁰C for 1 sec, the annealing process at 52⁰C for 1 min, the elongation process at 72⁰C for 2 min, and the post-processing process. Elongation at 72⁰C for 10 min. The amplification cycle starting from denaturation to elongation was repeated 40 times. FAdV PCR amplification using Forward Hexon (5'-CAARTTCAGRCAGACGGT-3') and Reverse Hexon (5'-TAGTGATGMCGSGACATCAT-3') primers [13]. A total of 10 µl of PCR products were analyzed by 2% agarose gel electrophoresis, and the gels were visualized under ultraviolet light. A positive test indicates a PCR product at the 897 bp (base pair) band [14].
DNA Sequencing
The purified product was mixed with Big Dye Terminator Kit version 3.1, RR-100 Big Dye Rerminator, 5x sequencing buffer, forward and reverses primers and NFW in a tube [15]. The contents in the tube were carefully homogenized and then transferred to a thermal cycler (ABI310 xL GENETIC ANALYZER, Applied Biosystem Inc.) programmed at 96°C for denaturation for 3 min. The cycle will be repeated 25 times and set at 96°C for 10 sec, 50°C for 5 sec, 60°C for 4 min. The sample was stored at 4°C until the sample was ready for deposition. Precipitation was carried out using 100% alcohol.
Homology Analysis
The sequenced nucleotide arrangement was then analyzed using the Needleman-Wuncsh Global Align Nucleotide Sequence contained in the BLAST program from the NCBI website [16]. This result will be used for the next process. The comparison isolate sample data obtained from Genbank was inputted into Bioedit Software ver 8.0 for alignment using ClustalW.
Phylogenetic Analysis
The origin data in the form of nucleotide and amino acid sequences from sample isolates and comparison isolates that were already in FASTA form were analyzed using ClustalW [17]. ClustalW results in the form of cladograms or phylogenetic trees can be read using treeviewsoftware. The phylogenetic tree was drawn using the maximum likelihood method contained in the MEGA ver 6.0 software.
RESULT
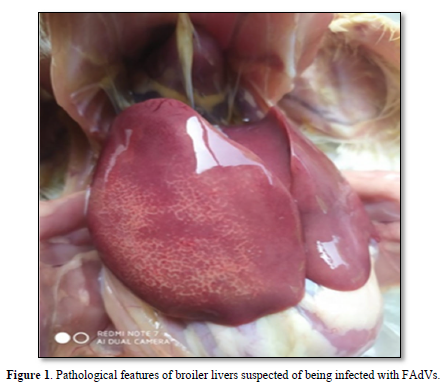
Samples were taken from broilers suspected to be infected with FAdVs and showed very clear anatomical pathological changes [18]. Figure 1 shows that the liver looks slightly swollen, striped, there are areas of pale, fatty, and brittle necrosis.
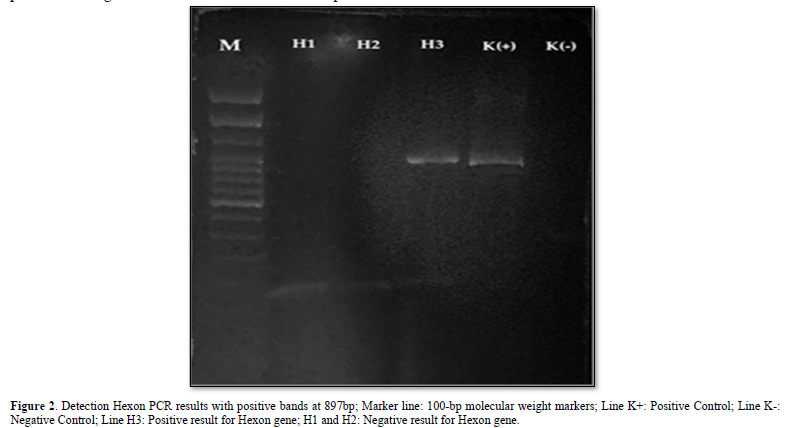
Samples were extracted by DNA extraction from the liver and PCR tests using Hexon forward and reverse primers [8], obtained PCR products from 3 samples, accompanied by positive and negative controls. The results of electrophoresis showed that there was only 1 sample that found DNA fragments; this was evidenced by the presence of a DNA band with a length of 897 bp as shown in Figure 2.
Sequencing results of IBH/CHK/H3/SLTG/2020 sample PCR products using the Applied Biosystem 3500xL Genetic Analyzer (BIONEER). The reference sequence used was isolate IBH/CHK/CHN/2018, Genbank: MG712775.1 and obtained different or changed nucleotides and fixed or the same, and can be seen in Figure 3.



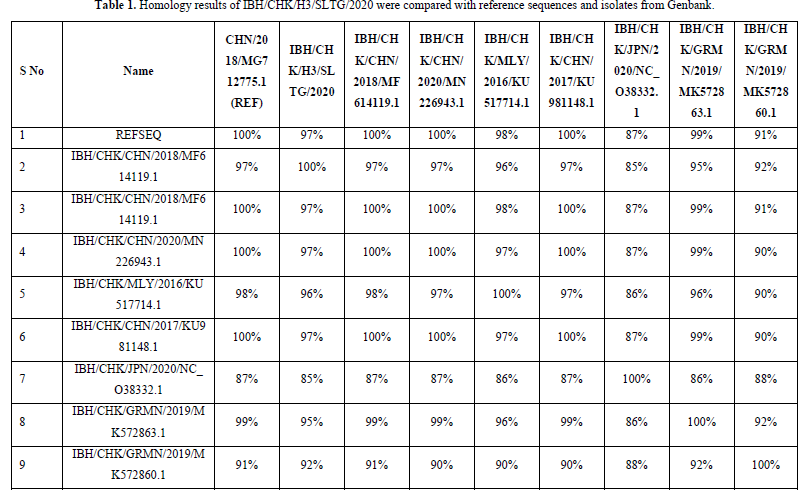
The results of the homology analysis of hexon protein sequences for the IBH/CHK/H3/SLTG/2020 samples were highest in the comparison of the IBH/CHK/H3/SLTG/2020 samples with the reference sequences of isolates MG712775.1 China/2018, MF614119.1 China/2018, MN226943. 1 China/2020 and KU981148.1 China 2017 show a homology of 97% as in Table 1, this indicates that there are many similarities in the sample nucleotide sequences with the comparison isolate above, and indicates that there is a similarity or approach of one species [19].
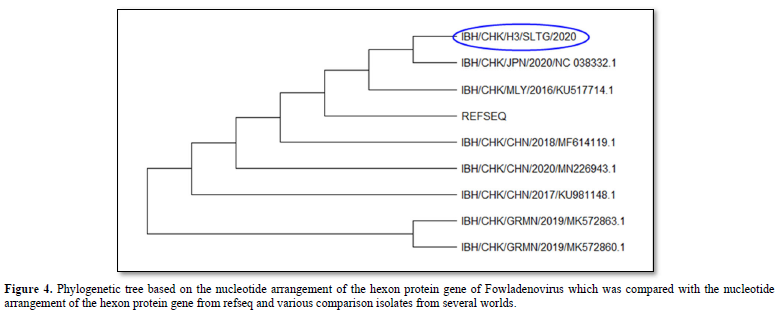
The results of the phylogenetic analysis showed that the IBH/CHK/H3/SLTG/2020 sample had a clade close to the reference sequence virus isolate - MG712775.1 China/2018, MF614119.1 China 2018, KU981148.1 China 2017, NC_038332 .1 Japan/2020 and MN226943.1 China 2020 as seen in Figure 4. Furthermore, these samples shared different clades with virus isolates MK5728631 Germany/2019, and MK5728601 Germany/2019.
DISCUSSION
This study used samples of liver organs from three broiler chickens from Salatiga which were found to have died suddenly and showed early clinical symptoms of IBH, namely the chickens looked lethargic, combs, wattles and facial skin looked pale, and at necropsy there was swelling of the liver. Degenerative changes and necrosis cause liver dysfunction which will affect blood osmotic pressure [20].
The results of hexon protein gene amplification from three samples of Fowladenovirus isolates found only one positive and two negative. The interpretation of the positive PCR results was due to the sample organ containing the target DNA [21]. While the interpretation of negative PCR results can be caused because the sample organ does not contain target DNA [21]. A negative result could also be due to the amount of virus present in the sample below the detection limit [22].
Nucleotide sequencing results obtained nucleotides that did not change or the same, and that changed or different. Genetic changes in one nucleotide base caused by mutations are usually associated with a large variety of viral genotypes [23].
The highest homology results were found in the comparison of IBH/CHK/H3/SLTG/2020 samples with reference sequence isolates MG712775.1 China/2018, MF614119.1 China/2018, MN226943.1 China/2020, KU981148.1 China 2017 showing a homology of 97 %, this indicates that there are many similarities or few differences between the nucleotide sequences of the IBH/CHK/H3/SLTG/2020 sample and the comparison isolate above [24]. The nucleotide sequence equation shows that there is a similarity or one-species approach between the sample isolates and the isolates in the above countries [25].
The results of the phylogenetic analysis showed that the IBH/CHK/H3/SLTG/2020 sample had a close clade with virus isolates with MG712775.1 China/2018 as the reference sequence, MF614119.1 China/2018, MN226943.1 China/2020, KU517714.1 Malaysia/2016, KU981148.1 China/2017, NC_038332.1 Japan/2020, MK572863.1 and different clade with virus isolate Germany/2019, MK572860.1 Germany/2019.
CONCLUSION
In conclusion, the presence of FAdVsinfectionin the IBH/CHK/H3/SLTG/2020 sample which has been tested using Polymerase Chain Reaction, is indicated by the formation of a DNA band along 897bp. The homology results show that the sample has a homology value of 97% with reference sequence isolates MG712775.1 China/2018, MF614119.1 China/2018, MN226943.1 China/2020, and KU981148.1 China 2017, and the results of the kinship relationship show that the sample IBH/CHK/H3/SLTG/2020 has a close clade with reference sequence virus isolates - MG712775.1 China/2018, MF614119.1 China 2018, KU981148.1 China 2017, NC_038332.1 Japan/2020 and MN226943.1 China 2020 and different clade with virus isolates MK572863.1 Germany/2019, and MK572860.1 Germany/2019.
ACKNOWLEDGEMENT
The author would like to express the gratitude for Department of Microbiology, Faculty of Veterinary Medicine, Airlangga University for the countless support.
No Files Found
Share Your Publication :